大家好,今天我们分享一个神奇的网站,提供各种可视化图片的代码和详解,生信小博士公众号内回复冒号后面的关键词领取网站地址:神奇网站
1.一R代码添加显著性
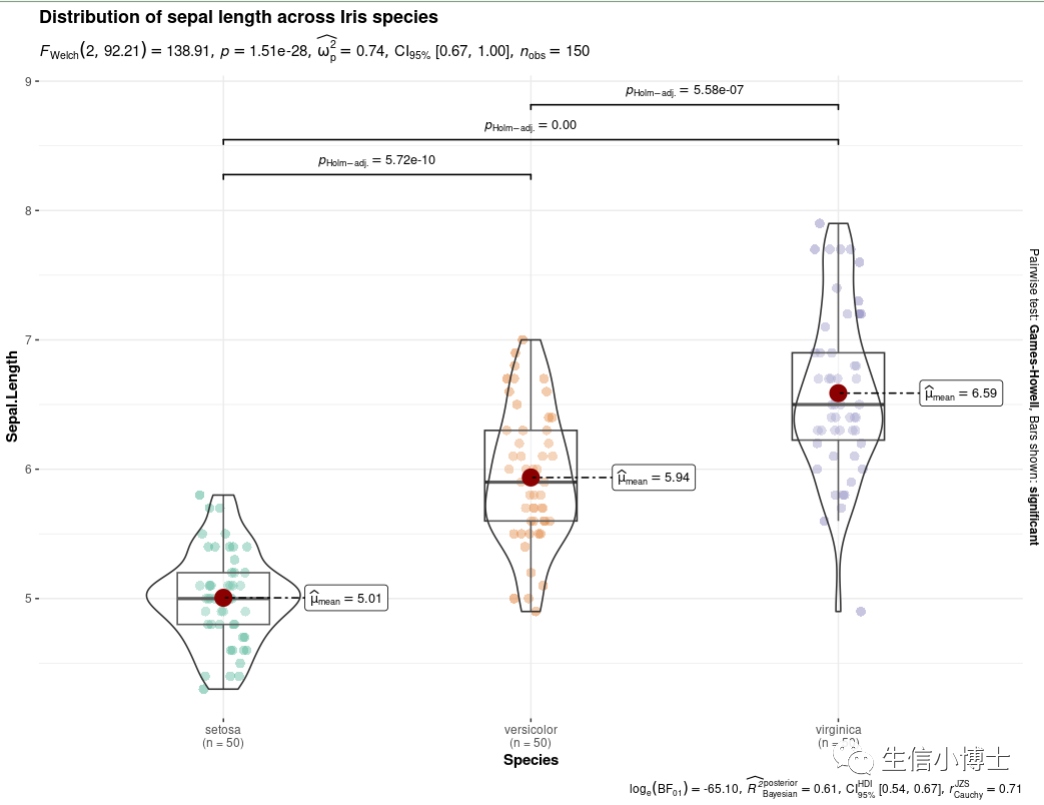
1#1----set.seed(123)library(ggplot2)library(ggstatsplot)ggbetweenstats(data = iris,x = Species,y = Sepal.Length,title = "Distribution of sepal length across Iris species")
还有详细参数的解释:
2.还可以换一种方式展示
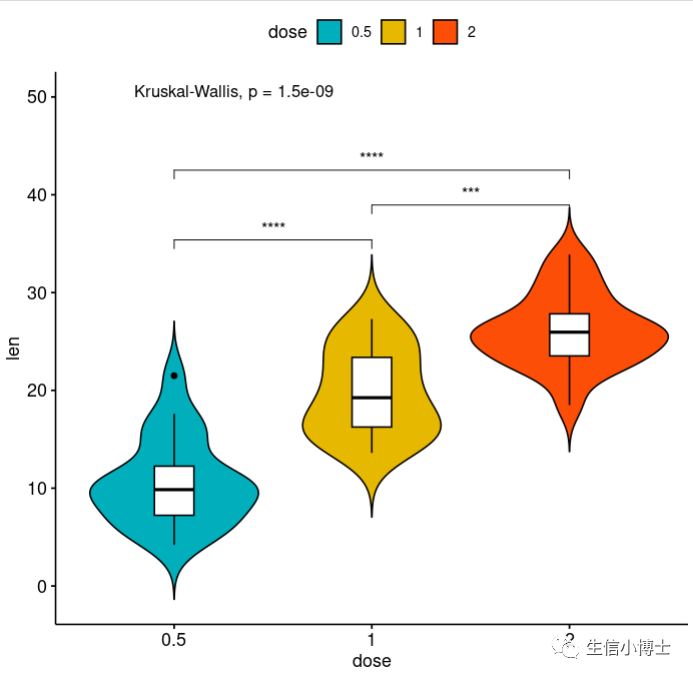
2#2------library(ggpubr)# Load datadata("ToothGrowth")df <- ToothGrowthhead(df, 4)# Add p-values comparing groups# Specify the comparisons you wantmy_comparisons <- list( c("0.5", "1"), c("1", "2"), c("0.5", "2") )ggboxplot(df, x = "dose", y = "len",color = "dose", palette =c("#00AFBB", "#E7B800", "#FC4E07"),add = "jitter", shape = "dose")+ stat_compare_means(comparisons = my_comparisons)+ # Add pairwise comparisons p-valuestat_compare_means(label.y = 50) # Add global p-value# Violin plots with box plots inside# :::::::::::::::::::::::::::::::::::::::::::::::::::# Change fill color by groups: dose# add boxplot with white fill colorggviolin(df, x = "dose", y = "len", fill = "dose",palette = c("#00AFBB", "#E7B800", "#FC4E07"),add = "boxplot", add.params = list(fill = "white"))+stat_compare_means(comparisons = my_comparisons, label = "p.signif")+ # Add significance levelsstat_compare_means(label.y = 50) # Add global the p-value
3. 单细胞的密度图
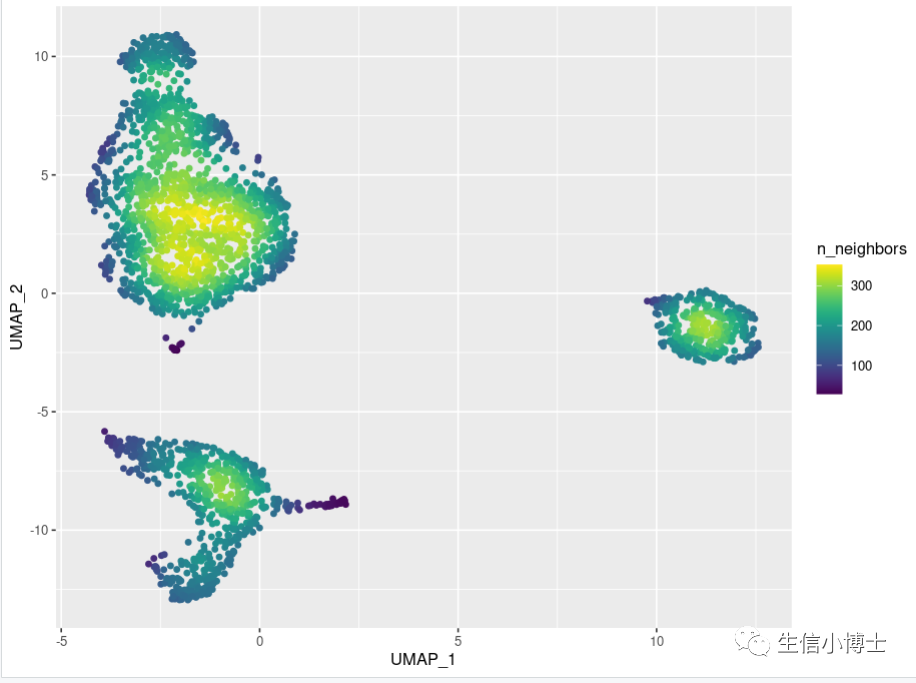
3# 3 单细胞密度图----pbmc=readRDS("~/gzh/pbmc3k_final.rds")library(Seurat)p=DimPlot(pbmc,label = TRUE)dat=p$datahead(dat)#install.packages("ggpointdensity")library(ggpointdensity)ggplot(data = dat, mapping = aes(x = UMAP_1, y = UMAP_2)) +geom_pointdensity(adjust = 4) +viridis:: scale_color_viridis()
4 生存曲线
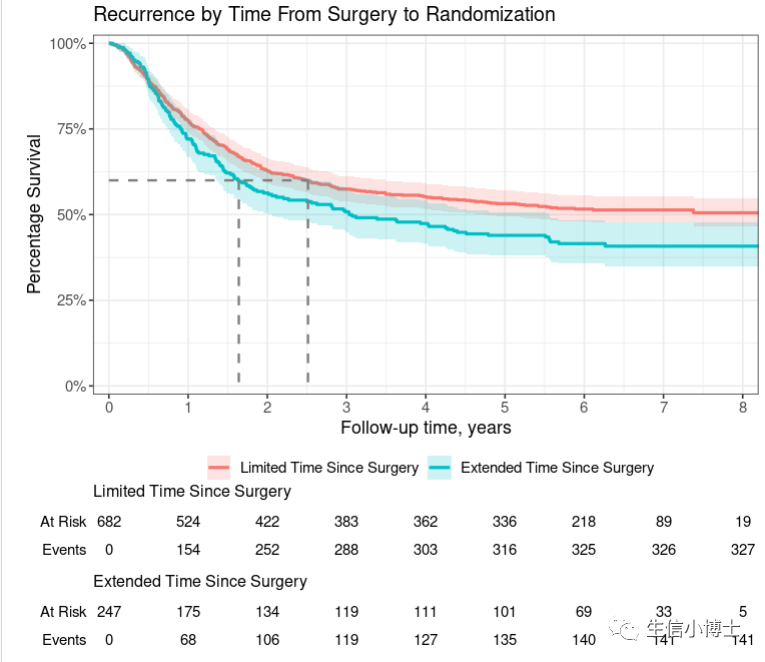
4 #4 生存曲线------#install.packages('ggsurvfit')library(ggsurvfit)#> Loading required package: ggplot2p <- survfit2(Surv(time, status) ~ surg, data = df_colon) |>ggsurvfit(linewidth = 1) +add_confidence_interval() +add_risktable() +add_quantile(y_value = 0.6, color = "gray50", linewidth = 0.75) +scale_ggsurvfit()pp +# limit plot to show 8 years and lesscoord_cartesian(xlim = c(0, 8)) +# update figure labels/titleslabs(y = "Percentage Survival",title = "Recurrence by Time From Surgery to Randomization",)



)



)

覆盖优化 - 附代码)


)






